
Eduardo Orias - 2/12/97
To go quickly to a particular major topic, click on the topic in the Table of Contents. At the end of each major topic section you may click to return to the Table of Contents or to the Main Page.
1. Tetrahymena thermophila The organism Nuclear dimorphism Life cycle 2. Micronuclear (germline) genetics Mendelian genetics Inducible conjugation variants Mapping genetically the MIC genome 3. Genetics of the macronucleus DNA rearrangements during macronuclear differentiation Phenotypic assortment Coassortment Mapping genetically and molecularly the MAC genome 4. Special topics of MAC genetics Sexual maturity Mating type differentiation Genetics of the ribosomal RNA gene (rDNA) MAC DNA replication and copy number control 5. Isolation and genetic characterization of mutants <to be added> 6. Other non-Mendelian inheritance Mitochondrial (Cytoplasmic) Genetics 10 Kb Plasmid Cortical Inheritance 7. On the evolution of nuclear dimorphism 8. Genetic Protocols and strategies <To be added> 9. Other Background Randomly Amplified Polymorphic DNA (RAPD) 10. References
*************************
*************************
Tetrahymena thermophila (Fig. 1) belongs to the Ciliated Protozoa-or Ciliates-a major, successful and diversified evolutionary lineage of unicellular eukaryotes. It is a freshwater organism that commonly inhabits streams, lakes and ponds. The cells are large: 40-50 um along the anterior-posterior axis. Like other ciliates, Tetrahymena cells have a striking variety of highly complex and specialized cell structures. Additional views of Tetrahymena cells can be found in the published literature: transmission EM [ref. 1, p. 5] and optical microscopy of stained cells [ref. 2, p. 180]. The Tetrahymena genome size (roughly 220 Megabase pairs) is of the same order of magnitude as that of Drosophila-one order larger than yeast (Saccharomyces) and one smaller than human.

************************
Tetrahymena's versatile biology provides extremely useful experimental tools. With a minimum doubling time below 2 hr, it is among the fastest growing eukaryotes. It readily grows to high density over a wide range of scale (from a few ul to fermentor scale) in a variety of media: totally defined synthetic chemical medium, rich nutrient broth or monobacterial culture. It grows over a wide temperature range: 12C to 41C. It has powerful active transport mechanisms and can be labeled with virtually any small molecule. The sexual stage in its life cycle, conjugation, can be induced at will, with high efficiency and synchrony. Cells can maintain DNA constructs introduced by transformation.
Tetrahymena is also a promising system for biotechnological applications. It is used for quick, reliable, sensitive and inexpensive bioassays. For example, it is used for determining the protein nutritional value of human foods, the toxicity of compounds of chemical or biological origin, and for monitoring water quality. It has potential for industrial synthesis of pharmaceuticals. Its capacity for massive, synchronous regulated secretion confers the potential for industrial synthesis of useful proteins. Since mosquitoes and snails are parasitized by other Tetrahymena species and closely related genera, T. thermophila has the potential for the development of tools for the biological control of major world-wide human diseases for which those organisms are intermediate vectors: schistosomiasis, malaria, yellow fever. Tetrahymena-based biotech strategies to fight fish hatchery and pet store epidemics of the closely related ciliate Ichthyophthirius ("Ick") are currently being explored.
This microbial eukaryote has great value as an experimental model system due to the facility with which it can be handled, its structural and functional differentiations, its accessibility to genetic and molecular approaches, its large evolutionary distance from other commonly used eukaryotic genetic model systems, and its potential for biotechnological applications coupled with its biosafety. It has provided an excellent system for the discovery and investigation of fundamental molecular and cellular mechanisms (e.g. ribozymes and telomeres). The availability of dozens of related species has proven extremely useful for identifying evolutionarily conserved domains in macromolecules and particularly for the dissection of important secondary structure and functional domains in RNAs that have or participate in catalytic activity. Its nuclear dimorphism provides Tetrahymena with an additional and unique dimension of biological versatility and genetic manipulability.
The species now called Tetrahymena thermophila was first considered to be Tetrahymena pyriformis. Later, T. pyriformis was found to comprise many sibling (cryptic) species, sexually isolated from one another but morphologically indistinguishable. As knowledge advanced, it was successively renamed variety 1 and syngen 1 of Tetrahymena pyriformis, before acquiring its current name [3]. Genetically, thermophila is the Tetrahymena species by far most extensively characterized.
Whenever possible I refer to useful reviews rather than primary research articles. Nanney's [2] introduction to the experimental biology of ciliates, with strong coverage of Tetrahymena, is particularly useful. A broad review of the biology of Tetrahymena was edited by Elliott [5]. A comprehensive review of ciliate molecular biology was edited by Gall [6]; it includes relevant chapters on conjugation, genetics, and DNA organization, all with heavy coverage of Tetrahymena thermophila. Prescott [4] comprehensive review of ciliate molecular genetics includes much about Tetrahymena. Bleyman [7] has recently reviewed the genetics of ciliates and loci determining interesting mutant phenotypes, including many in Tetrahymena. A shorter version of this article will be published as a chapter in ref. [16].
As is typical of ciliates, the nuclear apparatus of Tetrahymena is composed of two structurally and functionally differentiated types of nuclei, a phenomenon known as nuclear dimorphism. The micronucleus (MIC) is the germline, i.e. the store of genetic information for the sexual progeny. It is diploid and contains 5 pairs of chromosomes. No known genes are expressed in the MIC. Amicronucleate Tetrahymena cells (i.e., cells lacking a MIC) are frequently collected in nature, but in laboratory strains of T. thermophila the loss of the MIC leads to clone death. Only one viable laboratory-obtained amicronucleate cell line has been described. At cell division the MIC divides mitotically with kinetochores and intranuclear mitotic spindle.
The macronucleus (MAC) is the somatic nucleus, i.e. the nucleus actively expressed during vegetative multiplication. No known MAC DNA is transmitted to the sexual progeny. The MAC contains 200-300 autonomously replicating species derived from the 5 MIC chromosomes by site-specific fragmentation (further described below). The bulk of these DNA species is present at the average level of 45 copies per MAC. There are no visible structures expected for mitotic distribution of MAC pieces , such as kinetochores or mitotic spindle. The MAC is thus said to divide by amitosis. Approximately (but seldom exactly) half of the MAC DNA is distributed to each daughter MAC at cell division. Alternative allele copies of a locus segregate at random during MAC division (described later). Physical methods are available to preparatively separate and purify MICs and MACs from one another.
The life cycle consists of an alternation of haploid and diploid stages (haplophase and diplophase, respectively) with reference to the germline. Cell reproduction is exclusively by binary fission; it is exclusively asexual and occurs only in the diplophase. It is remarkable that such highly differentiated cells can divide by binary fission [e.g., see ref. 2, Plate II, p. 32]. Cell division is accompanied by a variety of morphogenetic events that result in the development of duplicate sets of cell structures, one for each daughter.
Conjugation is the sexual stage of the Tetrahymena life cycle, which includes no cell reproduction. During conjugation, two cells pair, form a temporary junction, exchange gamete nuclei and generate and differentiate the nuclear apparatus of their sexual progeny. The nuclear events of conjugation (Fig. 2) normally include meiosis, gamete nucleus formation, fertilization and nuclear differentiation. It is remarkable that at the time of exconjugant separation (stage 7 in Fig. 2) there are five nuclei experiencing four extremely diverse fates, all within a common cytoplasm. Conjugation includes the only-and very brief-haploid stage of the life cycle; it follows meiosis and quickly ends at fertilization. The haplophase is limited to a single nuclear division, without any cell division.
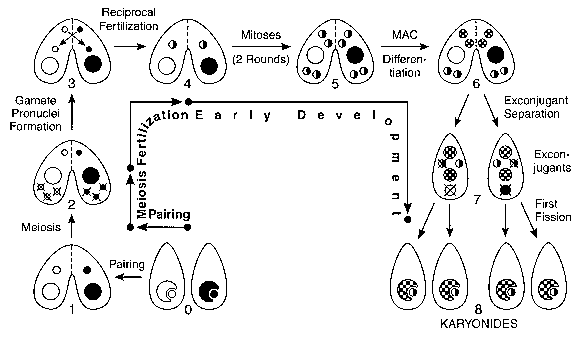
0: Vegetative cells homozygous for alternative alleles at one locus. The MIC (small circles) is shown nested in-but physically separate from-the MAC (large circles) , the normal arrangement in non-dividing vegetative cells. 1: Paired cells. 2: MICs undergo meiosis, and 4 haploid nuclei are produced. Only the anterior meiotic product remains functional; the other 3 disintegrate. This is the stage at which meiotic crossing-over, used for making genetic maps of the MIC genome, occurs. 3: Mitotic division of functional meiotic product yields genetically identical migratory (anterior) and stationary (posterior) gamete pronuclei. 4: Migratory pronuclei are reciprocally exchanged, and fuse with stationary pronuclei of the recipient cell, forming the zygote nucleus. 5: The zygote nucleus undergoes two mitotic divisions, giving rise to 4 genetically identical diploid nuclei. 6: Anterior products differentiate into macronuclei, while posterior products remain diploid micronuclei. This is the stage at which site-specific DNA rearrangements and mating type determination (discussed in the text) occur in the MAC. 7: Exconjugants separate. The old macronucleus, and one of the two new micronuclei, are destroyed. 8: Exconjugants undergo the first postzygotic cell division, forming the 4 karyonide cells. Each karyonide receives an independently differentiated new macronucleus, and a mitotic copy of the functional micronucleus. Karyonides then begin vegetative multiplication by binary fission.
************************
In order to conjugate, Tetrahymena cells must satisfy the following requirements: a) they must be starved for at least one required nutrient; b) they must be of different mating type; and c) they must have reached a sufficient level of sexual maturity. Seven mating types (I-VII) have been known since the earliest investigations of T. thermophila; the number has not increased after testing thousands of additional independent isolates from the wild. Sexual progeny normally are unable to mate again immediately after finishing conjugation. After 50-80 fissions they reach adolescence, i.e., they can mate with mature but not adolescent cells. Cells reach full sexual maturity 20-25 fissions later [8]. In the wild, the frequencies of the seven mating types tend to be equal and conjugation seems to occur frequently [9].
It is virtually certain that T. thermophila is an outbreeder in nature, i.e. with little or no mating among close relatives. Nevertheless, inbred strains have been successfully developed in the laboratory. Unlike Paramecium, there is no programmed somatic (vegetative) senescence and death: Tetrahymena cell lines are in practice immortal. The MIC of a cell line tends to lose chromosomes at a variable and unpredictable rate (germinal senescence). The lack of MIC gene expression prevents direct selection against such loss, just as in multicellular organisms possessing differentiated germline and soma. A useful (and still up-to-date) discussion of these topics is contained in Chapter. 9 of ref. [2].
Tetrahymena (and most Ciliates) are remarkably advanced among unicellular eukaryotes: no cell reproduction in the haplophase; differentiation of somatic and germ line nuclei; binary fission in the face of highly specialized compound cell structures; conjugation in diplophase; and internal fertilization at conjugation. Given this enormous biological versatility of ciliates, it is puzzling to this author why this evolutionary group includes no known multicellular forms.
Return to Table of Contents <<<<<<000>>>>>> Return to Main Page
The nuclear events of conjugation (Fig. 2) have two noteworthy genetic consequences: 1) they generate Mendelian genetics, as the diploid MIC undergoes meiosis and the zygote nuclei (from which progeny MIC and MAC are derived) are formed by the fusion of two haploid gamete nuclei. 2) The MIC and MAC of a karyonide start out genetically identical to one another; normal progeny are thus said to be homokaryons. 3) The differentiated MICs and MACs of the entire set of four karyonides derived from a given conjugating pair all have genetically identical diploid progenitors, and thus the four karyonides are expected to have identical phenotypes, no matter how different the parental cells were. It follows that pairs-and not exconjugants or karyonides-are the units to be counted in determining phenotypic ratios among the progeny of a cross.
There are several useful and well characterized variants of the normal conjugation pathway. Under some circumstances, conjugation is aborted and the differentiation of new MAC does not occur. The old MAC is retained instead. MAC retention can be readily identified (and excluded from progeny phenotypic ratio calculations) because the exconjugants remain sexually mature and continue to express parental phenotypes, including mating type. MAC retention is one way to generate heterokaryons, i.e., cells with genetically different MIC and MAC. MAC retention can be induced in every conjugating pair in crosses to star strains (genomic exclusion; see below). Heterokaryons are useful a) for positively selecting true progeny of conjugation in mass culture (e.g. parental cells with drug resistant MIC and sensitive MAC), b) for perpetuating lethal genotypes in the MIC (e.g., nullisomy, explained below) and c) for sorting out the contributions of old and new MACs toward the execution of early developmental events.
With low frequency, three cells can conjugate with one another in a mixture of cells of two mating types. Triplet conjugation usually generates either a set of three diploid exconjugants or a haploid-diploid-triploid set, depending on how well the three junctions are developed [see Chapter 7 in ref. 2]. If all three junctions are well developed, the triplet is symmetrical and each cell behaves equivalently: each conjugant donates a migratory pronucleus to the conjugant on its right, and receives one from the conjugant on its left. Three diploid exconjugants result, which generally are not genetically identical. If the junction between the two cells of like mating type is not well developed, then no pronucleus crosses this junction. The conjugant with two good junctions donates a migratory pronucleus to the conjugant on its right and receives a migratory pronucleus from the two other conjugants, becoming triploid. The other two exconjugants become diploid and haploid, respectively. The first well characterized nullisomic strains, i.e., strains lacking one or more pairs of chromosomes in the MIC, were obtained after meiosis of a haploid MIC within cells derived from a conjugating triplet.
Other useful and well characterized conjugation variants that occur spontaneously and can be induced at will are: genomic exclusion, cytogamy (i.e. self-fertilization), uniparental cytogamy and pronuclear fusion failure [see refs. 10-12]. Some of these variant conjugation pathways generate cells that are homozygous for their entire genome, either in the MIC only (genomic exclusion) or in both MICs and MACs (cytogamy and fusion failure). Cytogamy generates homokaryons, while fusion failure generates both hetero- and homokaryons. Cytogamy is used to efficiently isolate laboratory-induced recessive mutants. Pronuclear fusion failure is used for the generation of strains that are homozygous and simultaneously heterokaryons for several loci.
Genomic exclusion is induced by conjugation with a star strain. Such strains have a grossly defective MIC, with only a few percent of the normal DNA amount. The name genomic exclusion refers to the inability of the star strain to contribute any genetic information to the sexual progeny. When a normal cell conjugates with a star cell (i.e. round I of genomic exclusion), the prezygotic nuclear events occur normally in the normal conjugant. In the star conjugant, meiosis is initiated but no meiotic products are generated. The normal conjugant transfers its migratory pronucleus to the star conjugant, but gets nothing in return. Each conjugant thus ends up with a mitotic copy of the single meiotic product saved in the normal conjugant. This pronucleus diploidizes and becomes the MIC of the exconjugants, while their old MAC is retained. The diploidization of the MIC generates exconjugants that in one step have become homozygotes for their entire MIC genome and, depending on their initial genotype, often are heterokaryons. The star-derived exconjugant (but not the other exconjugant) retains the inability to maintain a normal MIC during subsequent asexual multiplication and soon reverts to the star state. The generation of homozygous heterokaryons is the most common application of genomic exclusion often in combination with phenotypic assortment (described later). Genomic exclusion involving a haploid strain as the normal conjugant was used to recover the first nullisomic strains.
Since round I exconjugants in genomic exclusion retain their old MACs, they retain their (different) mating types and are sexually mature. If left alone and not re-fed, they immediately undergo a second round of conjugation (round II of genomic exclusion). This is now a normal round of conjugation. If the two exconjugants from the same round I pair undergo round II, homokaryons with both MIC and MAC homozygous for the entire genome result. Ref. [11] contains an excellent illustration of genomic exclusion.
Additional methods to obtain whole-genome homozygotes involve induced blocks of fertilization. Cytogamy is a self-fertilization that occurs when the exchange of gamete pronuclei is blocked (e.g., by hyperosmotic shock or microtubule inhibitors). The resulting diploid new MIC is homozygous for the entire haploid genome of the functional meiotic product saved in each conjugant. Nuclear differentiation occurs normally and homokaryons are produced. Generally, the two exconjugants are genetically different. Uniparental cytogamy combines the induction of cytogamy with a first round of genomic exclusion. The resulting progeny are whole-genome homozygotes. In contrast to ordinary genomic exclusion, the first round exconjugants differentiate new macronuclei. The exconjugant clones mature sexually very early, a fortuitous circumstance that has been used to isolate mutants affecting the conjugation pathway [12].
Pronuclear fusion failure occurs when exchange of gamete pronuclei occurs but their fusion is blocked (e.g. with a pulse of microtubule inhibitors). Each conjugant at fertilization ends up with two nuclei, each of which is diploid and homozygous for an entire genome. Since each genome is derived from a different conjugant, the two nuclei are not necessarily genetically identical. Nuclear differentiation proceeds normally. Of the four new nuclei differentiated in each conjugant, one MIC and one MAC is derived from each of the two unfused pronuclei. Since one new MIC is destroyed, half of the karyonides are heterokaryons [10].
Genetic differences in Tetrahymena are mapped to chromosome arms by using nullisomic strains. These are strains lacking both copies of a chromosome or chromosome arm in their MIC. They survive because they are heterokaryons with a normal MAC. When a diploid strain homozygous for a recessive mutation is crossed to a nullisomic strain, the resulting monosomic progeny express the recessive genotype if the chromosome carrying the locus in question is absent from the nullisomic parent [illustrated in ref. 13 for mapping RAPD DNA polymorphisms]. By doing parallel crosses of the mutant to an appropriate set of nullisomics, the locus can be assigned to a chromosome arm. Bruns [14] and this web site list the chromosome arm assignments of many conventional loci, cloned DNA sequences and DNA polymorphisms. Parenthetically, the newly differentiated monosomic MAC starts out in a state of major gene dosage unbalance. While in most eukaryotes this condition would be lethal, Tetrahymena cells monosomic even for several chromosomes survive to be useful and reach normal growth rate because of mechanisms that regulate the copy number of MAC pieces during asexual multiplication.
As in other eukaryotes, genetic linkage maps of loci in individual chromosomes are constructed by exploiting the fact that meiotic crossing over generates recombinant genotypes with a frequency which, in the linear portion of the range, is roughly proportional to physical distance [see any genetics textbook, e.g. Chapter 5, ref. 15]. Major progress has recently been made in constructing a solid framework for a genetic map of the Tetrahymena MIC genome, included in this web site. This framework is currently based on the results of testing for linkage nearly 400 "randomly amplified polymorphic DNA" (RAPD) segments as well as some "classical" Tetrahymena genetic loci.
Return to Table of Contents <<<<<<000>>>>>> Return to Main Page
Several types of developmentally programmed DNA rearrangements occur during MAC differentiation, diagrammed in Fig. 3 and reviewed in [4]. One type is the site-specific fragmentation of the 5 MIC chromosomes into 200-300 subchromosomal molecular DNA species, here called MAC ARPs (autonomously replicating pieces). They have also been called minichromosomes. The average size of these pieces is roughly 700 Kb, and they range in size from a few hundred to a few thousand Kb; thus each ARP must contain many genes. The rDNA ARP (described later) is exceptionally small (21 Kb) and contains only the gene for the rRNA 45S precursor as an inverted repeat.

Top line: Arbitrary segment of MIC DNA. Thick bar: MAC-destined DNA segments. Open segments: internally deleted (MIC limited) DNA sequences. Thin connector: DNA sequences (including Cbs site-vertical arrow), lost during chromosome fragmentation. Second line: MAC ARPs derived from MIC chromosome segment. Wavy line: de novo added telomeric repeats. Dashed lines: boundaries of MIC-limited sequences. Bottom lines: Result of amplification in newly differentiated MAC. MAC chromosomes (averaging roughly 700 kb) are not drawn at the same scale as MIC limited sequences (generally in the order of 1 kb) or telomeric repeats (hundreds of bp).
************************
The Cbs (Chromosome breakage sequence) is a unique DNA 15-mer (5'AAAGAGGTTGGTTTA3' in one strand), necessary and sufficient for chromosome fragmentation during MAC differentiation. Telomeric repeats (GGGGTT at the 3' end) are added by telomerase at the newly created ends of these pieces, which are then amplified to the 45-ploid level; the rDNA, with 10,000 copies per MAC, is the only known exception. The Tetrahymena telomerase is a reverse transcriptase that includes the RNA template for telomeric sequence synthesis as an integral component of the enzyme. Telomeres are hundreds of bp long, and the number of GGGGTT/CCCCAA repeats varies from telomere to telomere. Tetrahymena cells with certain mutant telomere sequences senesce and die. The very high copy number of the rDNA telomere and the intensive telomere synthesis occurring during MAC differentiation made Tetrahymena a favorable system for the discovery of the molecular basis of eukaryotic telomeres and of telomerase, and for their continued investigation. The regulation of telomere length has important implications for carcinogenesis in mammalian cells [reviewed in ref. 17].
A seemingly unrelated type of site-specific rearrangement involves the deletion of internal DNA segments from the MAC during its differentiation. These are known as MIC-limited sequences or internally eliminated sequences (IES) [reviewed in ref. 18]. There are roughly 6,000 different deletion sites per haploid genome. At least a quarter of them are estimated to have alternative deletion ends. The alternative ends may be accurately specified or may loosely fall within domains of the order of 100 bp. Some imprecision may be tolerated in Tetrahymena because none of the deletion systems so far characterized have ends within protein coding sequences. The deletion size generally ranges from a few hundred to a few thousand bp. In total, roughly 15% of the MIC-derived DNA sequences are lost during MAC differentiation. MIC-limited sequences are AT-rich DNA. So far they have no known function and are proposed to be remnants of ancient invasions and dispersions of transposons.
When cells with a MAC initially heterozygous at a given locus undergo asexual multiplication, subclones that irreversibly express phenotypes associated with either homozygote are generated. Because recessive phenotypes come to expression in this way, the phenomenon was termed phenotypic assortment. Starting with a heterozygous cell with a mixed MAC, the steady state rate at which subclones pure for either allele arise is 0.011/fission. Assortment is attributed to the random distribution of allelic copies in a compound MAC (Fig. 4; see also ref. 2). Mathematically, the steady state rate of assortment of pure MACs from mixed MACs approaches 1/2N-1 per fission for a large N, where N is the number of copies just after MAC division (G1 stage). The measured rate led to the first determination of 45 as the average G1 ploidy of the MAC. This ploidy was subsequently confirmed by molecular measurements of MIC and MAC DNA amount and sequence complexity. When first described, the assorting units were believed to be diploid chromosome sets (subnuclei) showing allelic exclusion, i.e. with only one allele of the pair being expressed in each locus in each diploid set [see useful discussion in Chapter. 8 of ref. 2]. With increased knowledge about the molecular structure of the MAC, phenotypic assortment is now attributed simply to the random distribution of the acentromeric ARP copies at MAC division (Fig. 4). Phenotypic assortment has some similarities to the segregation of multicopy incompatible plasmids in bacterial clones.
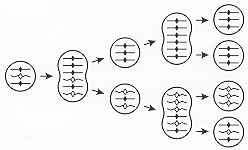
Only three of the 45 ARP copies are shown for simplicity. Circles: G1 MACs; "peanut-shells": amitotically-dividing MACs: each ARP copy has been replicated and its two copies have equal probability of going to the same or to different daughter MAC. Straight lines and solid loci symbols (diamonds): DNA from one parental cell; wavy lines and open symbols: DNA from the other parental cell. The diagram illustrates how a cell with a mixed MAC generates, through random distribution at successive MAC divisions, vegetative descendants that are pure for one allele or the other (phenotypic assortment). If one allele is dominant (e.g. solid diamonds), the initial cell expresses the dominant phenotype; later assortants pure for open diamonds, which express the recessive phenotype, arise. Please note: 1) At the first MAC division, the two wavy copies could, with equal probability, have been segregated to the two daughter MACs. Thus the rate of assortment of pure MACs with 3 ARP copies is 0.25 per fission. With 45 ARP copies, the steady state rate assortment of pure MACs is merely slower --0.011 per fission. 2) While each ARP copy is shown here to double exactly at each S period and the daughter MACs are shown here to receive exactly the same number of copies, such precision is probably rare in vivo in Tetrahymena MACs with 45 copies at G1.
************************
Phenotypic assortment allows any recessive allele to come to full expression. It also allows even a single mutant allele, generated by mutation or by DNA-mediated transformation, to replace completely the 45 wild type alleles in the MAC. Independent assortment, in combination with site-specific fragmentation, ensures extensive hereditary phenotypic diversification of the members of a vegetative clone, a situation commonly expected for Tetrahymena in the wild given the multiple heterozygosis predicted in an outbreeding species. Phenotypic assortment thus provides a second, somatic, shorter-term level of natural selection and adaptation for the species.
When doubly heterozygous cell lines are independently cultured asexually for hundreds of fissions, they exhibit virtually identical assortment pattern for certain pairwise combinations of neighboring loci [19]. In other words, starting with an AB/ab MAC, most of the vegetative descendants end up pure for either the AB or the ab (parental) combinations (Fig. 5). Less than 10% of the descendants become pure for either of the recombinant types (Ab or aB). This phenomenon is termed coassortment and allows the identification of coassortment groups, i.e. groups of loci such that its members all coassort with one another. A coassortment group is the MAC analog of a MIC meiotic linkage group. ARPs are the physical basis of coassortment groups; i.e., loci carried on the same ARP coassort, while loci carried on different ARPs assort independently. Coassortment makes it possible to map loci to MAC ARPs and determine ARP boundaries in the MAC purely by genetic means. Finding the physical ARP that carries a conventional locus can now be done indirectly by detecting its genetic coassortment with a physically mapped DNA polymorphism. These features may in the future facilitate the cloning of novel mutant genes in Tetrahymena that can only be cloned by complementation or chromosome walking.

A: Independent assortment of loci on two separate ARPs. B: Coassortment of loci on same ARP. Circular figure: MIC; "helmet"-shaped figure: MAC. Two different MAC ARPs (long and short) and three different loci (circles, triangles and diamonds) are shown. Only 8 of the 45 copies of each ARP are shown. The short ARP is omitted in panel B, as it is not needed for illustrating coassortment. Straight lines and solid loci symbols: DNA from one parental cell; wavy lines and open symbols: DNA from the other parental cell. Both parental cells are double homozygotes. P1 and P2: terminal assortants with parental genotypes; R1 and R2: terminal assortants with recombinant genotypes. Note that 1) Phenotypic assortment generated terminal assortants with MACs that are pure at each locus. 2) Coassortment between two loci is defined by a strong statistical excess of parental over recombinant types, not seen for independently assorting loci. 3) Loci on different MIC chromosomes (not shown here) are expected always to assort independently. 4) There is no assortment in the mitotically-dividing MIC, which remains doubly heterozygous.
************************
Genetic mapping of the MAC on the basis of coassortment is now underway. Currently identified coassortment groups are listed in this web site. The most carefully mapped coassortment groups so far represent contiguous segments of MIC DNA, as expected from the simple model shown in Fig. 3. A few cloned DNA segments have been physically assigned to ARPs by hybridizing labeled probe to Southern blots of whole cell DNA separated in agarose gels by pulsed field electrophoresis [illustrated in ref. 19].
MIC-limited polymorphic DNA sequences are expected to show no assortment during vegetative multiplication because the signal originates in the MIC, which divides mitotically and remains heterozygous. This lack of assortment has been used to detect potential MIC-limited RAPD polymorphisms. Their status has been confirmed by PCR or Southern blot analysis: the signal disappears when whole cell DNA from a nullisomic strain missing the chromosome that carries the MIC-limited sequence is tested [unpublished observations, Orias lab]. A list of MIC-limited RAPDs and their MIC map location is given in this web site. Not all apparent lack of assortment is due to MIC-limited DNA. Exclusive assortment to a particular allele can result from differential replication/maintenance of the ARP copy carrying that allele [21]. For certain natural genetic polymorphisms in surface antigen (serotype), extremely biased initial amplification of one particular allele (in the absence of differential vegetative replication) is sufficient to explain the observed appearance of lack of assortment [27]
Return to Table of Contents <<<<<<000>>>>>> Return to Main Page
Sexual maturation is a stable, somatically inherited differentiation of the MAC, remarkable in that it must occur in response to some endogenous fission counting program, as these are free-living unicells. The differentiation of new MACs under conditions of physiological stress induces early maturity.
The MIC carries the potential for 5-7 mating types-depending on the genotype at the mating type (mat) locus. Nevertheless, a cell generally expresses only one mating type. This is the result of a somatically inherited, irreversible, stochastic event that occurs in the differentiating new macronucleus. The frequencies with which various mating types arise are not necessarily equal, and are affected by environmental conditions prevailing at the time of MAC differentiation, e.g. temperature and starvation [see ref. 20].
Roughly 50% of newly differentiated MACs normally are pure for determinants for a single mating type. The rest of the new MACs are mixed but, as a result of phenotypic assortment, most MACs have become pure for a single mating type determinant by the time sexual maturity is reached. Sexually mature cells with MACs that remain mixed give rise to clones within which pairing can occur; they are called selfers. Continued asexual multiplication of selfers generates subclones with pure MACs. The genetics of selfers was the unusual context within which phenotypic assortment was first described and analyzed in Tetrahymena. A model of mating type determination based on an alternative deletion system has been proposed for T. thermophila [20]. It explains quantitatively the genetic phenomenology of mating type determination and predicts the order of mating type genes within the mat locus, but remains to be tested molecularly.
Tetrahymena is an exceptional organism in having only a single copy (per haploid genome) of the gene for the 45S ribosomal RNA precursor, which is processed posttranscriptionally to the 18S and 28S rRNAs. During MAC differentiation, this copy is excised by Cbs-directed fragmentation, turned into a 21 Kb inverted repeat (palindrome), supplied with telomeres and amplified to the level of roughly 10,000 copies per MAC [illustrated in ref. 21]. All but the central 29 bp of the rDNA is palindromic. The two halves of the palindrome are transcribed divergently. Each half consists of a central transcribed region, flanked by the 5'- and the 3'-nontranscribed spacers (NTS). The 5'-NTS contains the origin of rDNA replication. Because of its high copy number, the rDNA telomere represents roughly half of the telomeres in the MAC. Tetrahymena rDNA telomeres are built into the vector used to clone large DNA inserts in Saccharomyces as Yeast Artificial Chromosomes (YACs). The 28S rRNA coding sequence includes an intron; understanding of its self-splicing nature led to the codiscovery of ribozymes [22]. Paromomycin resistance is a useful selectable trait, determined by a mutation located near the 3'-end of the 16S rRNA coding region. Most current Tetrahymena transformation vectors incorporate the rDNA origin of replication, which is required for their maintenance in transformants.
Heterozygotes for Pmr assort to PM-S with kinetics that correspond roughly to that of 45 randomly distributed (G1) copies [28]. This number is much smaller than the total number of rDNA copies (roughly 10,000) or the number of nucleoli (roughly 500 in exponentially growing cells). Any strong, systematic tendency for sister rDNA molecules to go to the same daughter at MAC division would lower the apparent number of assorting units. In addition, the known differential growth of PM-S relative to PM-R cells would spuriously lower the apparent number of assorting units.
The rDNA is localized in nucleoli, of which there are about 500 in exponentially growing cells [illustrated in ref. 1, p. 5]. The origin of replication of the rDNA has been mapped to the 5'-nontranscribed spacer. A natural polymorphism and laboratory mutations that affect the relative replicative maintenance of the rDNA during asexual multiplication in heterozygotes have been described [reviewed in ref. 21]. These studies reveal a coupling between the initiation of rDNA replication and rRNA transcription. Inclusion of the 5'-nontranscribed spacer of the rDNA containing mutations that confer replicative advantage has proven to be very useful for insuring the maintenance of recombinant DNA constructs in high copy number in the MAC after introduction by transformation. Fortunately, the maintenance in high copy number of a plasmid carrying the rDNA replication origin is compatible with the maintenance of the endogenous rDNA also in high copy number.
Several observations imply the existence of mechanisms that regulate ARP copy number in the bulk of the MAC DNA. 1) The Tetrahymena MAC has no visible machinery to ensure the regular distribution of daughter copies of DNA molecules and the genetic evidence indicates that distribution of allele copies is random. 2) The distribution of total bulk DNA to daughter MACs is seldom equal. This imprecision predicts descendants with lethal DNA sequence deficiencies and imbalances in higher frequency than is actually observed. 3) Cells that start out with haploid, triploid or extensively aneuploid MACs generate descendants with the normal amount of MAC DNA. Two levels of regulation must be envisioned: a global mechanism that maintains every ARP (other than the rDNA) at an average of 45 copies, and a specific mechanism that corrects copy number deviations of each individual ARP independently of the deviations occurring at other ARPs. Unusual cell cycles (with reference to the MAC) are triggered in cells with a total G1 amount of MAC DNA outside a predetermined range. When the upper threshold is exceeded, the cell divides without MAC DNA replication; when the G1 amount is below the lower threshold, two rounds of MAC DNA replication occur before cell division. Virtually everything remains to be learned about the mechanisms that control replication and copy number in the Tetrahymena MAC.
Return to Table of Contents <<<<<<000>>>>>> Return to Main Page
Tetrahymena cells contain 600-800 mitochondria [see ref. 1, p. 75]. Their DNA is a linear DNA molecule, an uncommon occurrence among eukaryotic mitochondria. Its telomeres consist of a variable number of repeats of a 53-bp unique sequence. Chloramphenicol resistance, determined by a mutation virtually certain to reside in the mitochondria, shows cytoplasmic inheritance [see ref. 7]: i.e., the exconjugant clone derived from a resistant conjugant remains resistant, while the exconjugant clone derived from the sensitive parent remains sensitive. The genetic evidence suggests that in Tetrahymena the exchange of mitochondria across the conjugal junction occurs rarely, if ever.
A linear 10 Kb plasmid (pTtL10), present in roughly 20,000 copies per cell, has been described in certain wild isolates of T. thermophila [23]. Its ends are terminal inverted repeats of a 47 bp unique sequence. This organization-but not the repeat sequence itself-is reminiscent of the mitochondrial DNA telomeres. Its 5' (but not 3') ends are resistant to exonuclease digestion. This plasmid is suspected of having protein-primed (rather than RNA-primed) DNA replication, such as found in adenovirus, for example. When plasmid-free cells conjugate with plasmid-containing cells, the plasmid is transferred to the plasmid-free conjugant in some but not all the pairs. No plasmid transfer has been detected in mixed vegetative culture. The cellular location of the plasmid DNA may well be mitochondrial. It has been proposed that in the course of evolution the Tetrahymena mitochondrial DNA became linear as a consequence of recombination with a pTtL10-type plasmid.
The somatic ciliature in Tetrahymena is distributed throughout the cell and organized in a regular array of rows (or meridians) of ciliary units in the cell cortex [illustrated in ref. 2, p.180]. Each ciliary unit carries one cilium and associated cytoskeletal structures, including several microtubule arrays. The ciliary unit has anterior-posterior and left-right asymmetry. Variant cells having ciliary rows with reversed polarity can be generated in the absence of genic mutations; the reversed polarity is hereditarily transmitted at cell division. The (nongenic) basis of this form of inheritance (cortical inheritance or cytotaxis) is the highly structured addition of new ciliary units that compensates for their dilution at cell division. Overall orientation of a ciliary row is perpetuated through cell division because a) a new ciliary unit always differentiates anteriorly (i.e. along the meridian) and in tandem orientation with regard to the local organization of a preexisting ciliary unit and b) fission occurs in the equatorial plane [see useful in Chapter. 11 of ref. 2].
An unusual pattern of inheritance was observed when analyzing certain mutagenized progeny selected for the absence of expression of the immobilization (cell surface) antigen specified by the serH locus[29]. By analogy to well established genetic observations in Paramecium [30], it has been proposed that in these variants the altered cytoplasm in which a new MAC differentiates can hereditarily determine the state of expression of the serH locus in the asexual clone that carries descendants of that MAC. It has further been speculated that this phenomenon is mediated at the level of DNA rearrangements in the differentiating MAC. These experiments need to be repeated under conditions where independent genetic markers (now available) provide a rigorous check against unexpected cytogenetic accidents that could distort interpretations.
A remarkable phenomenon has been recently reported in Tetrahymena [31] which may provide a basis for the above observations. Cell lines can be obtained in which a certain MIC-limited sequence fails to be removed from the MAC. When a new MAC differentiates in the presence of such an old MAC, this condition is perpetuated, i.e. the normally MIC limited sequence is retained in the newly differentiated MAC. The basis for this observation remains to be elucidated, but the phenomenon appears at least superficially related to analogous observations made in species of the Paramecium aurelia group of ciliates [32].
The nuclear dimorphism of the ciliates is a remarkable phenomenon. It staggers the imagination to consider how amitosis of an acentromeric fragmented somatic genome could have evolved from mitosis through a series of functional intermediates. The following steps have been proposed for the evolution of the MAC. The first major step would have occurred in distant eukaryotic ancestors having an alternation of haploid and diploid phases in their life cycle. A differentiated, non-dividing somatic MAC could then have evolved in the context of a diplophase consisting exclusively of a non-dividing, unicellular trophic (feeding) stage [24]. This would have previously required the evolution of mechanisms that inhibit diplophase cell division and could well have been accompanied by the loss of mitotic mechanisms and components in the diplophase, in the absence of selection against such unnecessary mechanisms. Such a life cycle and type of nuclear dimorphism is found today among certain living Foraminifera [25], a group of shelled, marine unicellular eukaryotes.
The proposed second major step is the reacquisition of cell division in the diplophase-through events akin to cancer-causing mutations in multicellular organisms-and the gradual shortening of the haplophase, giving rise to Karyorelict ciliate type of nuclear dimorphism [24]. These ciliates have MACs that do not divide. The loss of the MAC during cell multiplication is averted by the fact that at every vegetative cell cycle the MIC divides twice; half of the products differentiate into MACs and are segregated to the two daughter cells (much like in Tetrahymena's first cell cycle after zygote formation; step 7-8 in Fig. 2). The maintenance of a non-dividing type of nucleus in a dividing cell is an unprecedented feature of Karyorelict ciliates, readily rationalized by this proposed evolutionary history. Absent nuclear division, chromosome fragmentation could have evolved at this or the previous stage without any risk of chromosomal imbalance caused by the lack of centromeres on the fragments. Ploidy increase may have happened at this stage to support the evolutionary increase in size of the cells.
The last major step would be the reacquisition of division in MACs with relatively high ploidy as a superior alternative to MAC differentiation at every cell cycle [26]. Absent mitotic mechanisms and functional centromeres in the MAC, the early evolutionary intermediates would have found irregular, serendipitous ways to distribute daughter DNA copies at MAC division. The possession of high ploidy in the MAC would have lessened the risks of chromosomal loss or serious imbalance associated with unequal distributions. The validity of these evolutionary speculations remains to be determined by future research.
Return to Table of Contents <<<<<<000>>>>>> Return to Main Page
The RAPD method (randomly-amplified polymorphic DNA) is an efficient method for detecting DNA polymorphisms [33]. This method uses 10-mers of arbitrary sequence as primers for PCR amplification. Up to roughly a dozen discrete ethidium bromide- stained bands are revealed with each primer. The pattern is specific for each primer, and is sensitive to single base changes in the primer; thus each band checks 20 base pairs for single (or more) base polymorphisms. By screening two different wild type or lab inbred strains the method uncovers in effect a practically inexhaustible source of "instant mutations".
The RAPD method appears to provide an efficient, general, relatively inexpensive and safe method to obtain genetic maps of complex genomes. RAPD polymorphisms have the additional advantage that the polymorphic DNA is molecularly cloned by the very first act of polymorphism detection. Minute amounts of template DNA are required (25 ng per reaction). Anyone can reproduce the polymorphism and generate a sample of the polymorphic DNA segment simply by using the corresponding 10-mers, which are commercially available at an inexpensive price. A list of the RAPD PCR primers used in mapping the Tetrahymena genome is given in this web site.
Special features of Tetrahymena genetics, uniquely related to their nuclear dimorphism, have further enhanced
the power of RAPD mapping [13,34].
1) Genomic exclusion allows the isolation of meiotic segregants as "instant" whole-genome homozygotes
from independent, single meiotic products of a multiple-heterozygous F1. This represents a far greater degree of
homozygosity than can be accomplished by ordinary selfing of an F1 individual. The direct generation of whole-genome
homozygotes is extremely valuable in view of the "dominance" of the band+ RAPD phenotype, i.e. the inability
of the RAPD method to distinguish between band+/band- heterozygotes and band+ homozygotes.
2) The availability of nullisomic strains has allowed us to assign linkage groups to particular MIC chromosome
arms, to target RAPD screens to particular chromosome segments and to identify MIC-limited RAPDs.
3) The ability to map RAPDs to MAC ARPs by coassortment provide an additional, experimentally independent check
on the maps generated by meiotic linkage.
Return to Table of Contents <<<<<<000>>>>>> Return to Main Page
1. Hill,DL (1972): The Biochemistry and Physiology of Tetrahymena. Academic Press, New York & London. 230 pages.
2. Nanney,DL (1980): Experimental Ciliatology: An introduction to genetic and developmental analysis in Ciliates. John Wiley & Sons, New York, NY.
3. Nanney,DL; McCoy,JW (1976): Characterization of the species of the Tetrahymena pyriformis complex. Trans. Amer. Microsc. Soc. 95, 664-682.
4. Prescott,DM (1994): The DNA of ciliated protozoa. Microbiol. Rev. 58, 233-267.
5. Elliott,AM (1973): Biology of Tetrahymena. Dowden, Hutchinson and Ross, Stroudsburg, PA. 508 pages.
6. Gall,JG (1986): Molecular Biology of the Ciliated Protozoa. Academic Press, New York, NY.
7. Bleyman,LK (1996): Ciliate genetics. In: Ciliates. Cells as Organisms. (Eds: Hausmann,K; Bradbury,PC) Gustav Fisher Verlag, Stuttgart - Jena - New York, 291-324.
8. Rogers,MB; Karrer,KM (1985): Adolescence in T. thermophila. Proc. Nat. Acad. Sci. USA 82, 436-439.
9. Doerder,FP; Gates,MA; Eberhardt,FP; Arslanyolu,M (1995): High frequency of sex and equal frequencies of mating types in natural populations of the ciliate Tetrahymena thermophila. Proc. Natl. Acad. Sci. U. S. A. 92, 8715-8718.
10. Orias,E (1986): Ciliate Conjugation. In: Molecular Biology of the Ciliated Protozoa. (Ed: Gall,JG) Academic Press, New York, NY, 45-84.
11. Bruns,PJ (1986): Genetic organization of Tetrahymena. In: The Molecular Biology of Ciliated Protozoa. (Ed: Gall,JG) Academic Press, New York, NY, 27-44.
12. Cole,ES; Bruns,PJ (1992): Uniparental cytogamy: a novel method for bringing micronuclear mutations of Tetrahymena into homozygous macronuclear expression with precocious sexual maturity. Genetics 132, 1017-1031.
13. Brickner,JH; Lynch,TJ; Zeilinger,D; Orias,E (1996): Identification, mapping and linkage analysis of randomly amplified DNA polymorphisms in Tetrahymena thermophila. Genetics 141, 1315-1325.
14. Bruns,PJ; Cassidy-Hanley,D (1993): Tetrahymena thermophila. In: Genetic Maps: Locus Maps of Complex Genomes, 6th ed. (Ed: O'Brien,SJ) Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2175-2179.
15. Griffiths,AJF; Miller,JH; Suzuki,DT; Lewontin,RC; Gelbart,WM (1996): An Introduction to Genetic Analysis. 6th ed. W.H.Freeman and Company, New York. 915 pages.
16. Reeve,ECR (1998): Encyclopedia of Genetics. Fitzroy Dearborn Publishers, London. 000 pages.
17. Greider,CW; Blackburn,EH (1996): Telomeres, telomerase and cancer. Sci. Am. 274, 92-97.
18. Yao,M-C (1996): Programmed DNA deletions in Tetrahymena: Mechanisms and implications. Trends in Genetics 12, 26-30.
19. Longcor,MA; Wickert,SA; Chau,M-F; Orias,E (1996): Coassortment of genetic loci during macronuclear division in Tetrahymena thermophila. Eur. J. Protistol. 32, 85-89.
20. Orias,E (1981): Probable somatic DNA rearrangements in mating type determination in Tetrahymena thermophila: A review and a model. Devel. Genet. 2, 185-202.
21. Kapler,GM (1993): Developmentally regulated processing and replication of the Tetrahymena rDNA minichromosome. Curr. Opin. Genet. Dev. 3, 730-735.
22. Kruger,K; Grabowski,PJ; Zaug,AJ; Sands,J; Gottschling,DE; Cech,TR (1982): Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence in Tetrahymena. Cell 31, 147-157.
23. Ortiz,JM (1993): Characterization of an Extrachromosomal Linear DNA molecule discovered in Tetrahymena. Ph.D. Dissertation, University of California at Santa Barbara, Santa Barbara, CA. 101 p.
24. Orias,E (1991): On the evolution of the karyorelict ciliate life cycle: heterophasic ciliates and the origin of ciliate binary fission. BioSystems 25, 67-73.
25. Grell,KG (1979): Cytogenetic systems and evolution in Foraminifera. J. Foraminif. Res. 9, 1-13.
26. Orias,E (1991): Evolution of amitosis of the ciliate macronucleus: Gain of the capacity to divide. J. Protozool. 38, 217-221.
27. Nanney,DL; Preparata,RM (1979): Genetic evidence concerning the structure of the Tetrahymena thermophila macronucleus. J. Protozool. 26, 2-9.
28. Merriam,EV; Bruns,PJ (1988): Phenotypic assortment in Tetrahymena thermophila: assortment kinetics of antibiotic-resistance markers, tsA, death, and the highly amplified rDNA locus. Genetics 120, 389-395.
29. Doerder,FP; Berkowitz,MS (1987): Nucleo-cytoplasmic interaction during macronuclear differentiation in ciliate protists: genetic basis for cytoplasmic control of SerH expression during macronuclear development in Tetrahymena thermophila. Genetics 117, 13-23.
30. Sonneborn,TM (1977): Genetics of cellular differentiation: stable nuclear differentiation in eucaryotic unicells. Ann. Rev. Genet. 11, 349-367.
31. Chalker,DL; Yao,MC (1996): Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol. Cell. Biol. 16, 3658-3667.
32. Meyer,E; Duharcourt,S (1996): Epigenetic regulation of programmed genomic rearrangements in Paramecium aurelia. J. Eukaryot. Microbiol. 43, 453-461.
33. Williams,JGK; Kubelik,AR; Livak,KJ; Rafalski,JA; Tingey,SV (1990): DNA polymorphisms amplified by arbitrary primers are useful genetic markers. Nucl. Acids Res. 18, 6531-6535.
34. Lynch,TJ; Brickner,JH; Nakano,KJ; Orias,E (1995): Genetic map of randomly amplified DNA polymorphisms closely linked to the mating type locus of {ITetrahymena thermophila}. Genetics 141, 1315-1325.
Return to Table of Contents <<<<<<000>>>>>> Return to Main Page